Borja Camino-Pontes, Marilyn Gatica, Antonio Jimenez-Marin, Patricio Orio, Rodrigo Cofre, Jesus M Cortes. Modular structure of high-order interactions in the human brain. OHBM 2022 – Organization for Human Brain Mapping [pdf]
Introduction:
A network is highly modular when different communities of nodes have high intra-connectivity within them and low inter-connectivity between them. Different methods and strategies have been used to maximize modularity in brain networks, see for instance [1] and references therein, resulting in a list indicating which node belongs to which community. Despite some strengths and weaknesses between the different methods, most of them start from a connectivity matrix that defines pairwise interactions between network nodes. Following previous work [2]–[4], we built here connectivity matrices defining node high-order interactions, from triplets to n-plets, and compared different communities obtained across different modularity methods.
Methods:
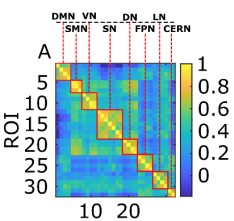
K = 86 healthy subjects from the Human Connectome Project (HCP), all of them were healthy unrelated subjects, 43 females, with a mean age of 28,28 years (σ=3,6 years). Resting-state fMRI data was pre-processed with the ICA-FIX pipeline provided by HCP. We extracted M = 32 region-level time-series of the different 8 resting-state networks (RSNs) incorporated into the CONN platform [5], namely, Default Model Network (DMN), Sensory-Motor Network (SMN), Visual Network (VN), Salience Network (SN), Dorsal Attention Network (DAN), Fronto-Parietal Network (FPN), Language Network (LN) and Cerebellear Network (CERN). We first calculated the O-information that accounts for high-order interactions in n-plets of regions [2], and particularized for subsequent analyses to n=3, i.e. the high-order interactions in triplets, also known as interaction information [6], [7]. If the value of the O-information is greater than 0, the interaction in the triplet is said to be redundant, and, if it is lower than 0, it is said to be synergetic. Next, we built connectivity matrices Cm for each value of m by defining, where λ is a free parameter and Rm and Sm are respectively the redundancy and the synergy in the triplet interaction between any two regions when interacting with region m. Notice that we have a different connectivity matrix Cm for each value of m. Eq. (1) allows to parametrize different nature of high-order interactions by tuning the parameter λ, eg., Cm = Sm for λ=0, Cm = Rm for λ=1, while competing redundant and synergetic interactions exist for λ values between 0 and 1. Finally, when considering each region m=1,…, M as one possible node in the network, and using different maximization algorithms, the modularity of different interacting networks Cm is maximized, e.g., using the Louvain algorithm, and the different resulting communities are discussed.
We also made use of different network partitions such as the Brain Hierarchical Atlas[8], Desikan[9] and Schaefer[10] .
Results:
The submitted figure illustrates different scenarios. For M=32 regions defined in the partition of the CONN RSNs, Fig 1A illustrates the pairwise functional connectivity matrix obtained for comparison purposes. Figs 1B-C show average across subjects and regions of the different matrices Rm and Sm. We have obtained different network communities in the different scenarios, comparing different methods and using different atlases. We also analysed a continued parameterization depending on λ, and assess the participation that different brain regions play across different communities.
Conclusions:
High-order interactions in the human brain reveal undetectable relations by pairwise-interaction strategies. The study assesses the modular structure of the space governing those high-order interactions.
References
[1] O. Sporns y R. F. Betzel, «Modular Brain Networks», Annu. Rev. Psychol., vol. 67, n.o 1, pp. 613-640, ene. 2016, doi: 10.1146/annurev-psych-122414-033634.
[2] F. E. Rosas, P. A. M. Mediano, M. Gastpar, y H. J. Jensen, «Quantifying high-order interdependencies via multivariate extensions of the mutual information», Phys. Rev. E, vol. 100, n.o 3, p. 032305, sep. 2019, doi: 10.1103/PhysRevE.100.032305.
[3] M. Gatica et al., «High-Order Interdependencies in the Aging Brain», Brain Connect., vol. 11, n.o 9, pp. 734-744, nov. 2021, doi: 10.1089/brain.2020.0982.
[4] F. Battiston et al., «Networks beyond pairwise interactions: Structure and dynamics», Phys. Rep., vol. 874, pp. 1-92, ago. 2020, doi: 10.1016/j.physrep.2020.05.004.
[5] S. Whitfield-Gabrieli y A. Nieto-Castanon, «Conn: A Functional Connectivity Toolbox for Correlated and Anticorrelated Brain Networks», Brain Connect., vol. 2, n.o 3, pp. 125-141, jun. 2012, doi: 10.1089/brain.2012.0073.
[6] A. Erramuzpe et al., «Identification of redundant and synergetic circuits in triplets of electrophysiological data», J. Neural Eng., vol. 12, n.o 6, p. 066007, dic. 2015, doi: 10.1088/1741-2560/12/6/066007.
[7] B. Camino-Pontes et al., «Interaction Information Along Lifespan of the Resting Brain Dynamics Reveals a Major Redundant Role of the Default Mode Network», Entropy, vol. 20, n.o 10, p. 742, sep. 2018, doi: 10.3390/e20100742.
[8] I. Diez et al., «A novel brain partition highlights the modular skeleton shared by structure and function», Sci. Rep., vol. 5, n.o 1, p. 10532, sep. 2015, doi: 10.1038/srep10532.
[9] R. S. Desikan et al., «An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest», NeuroImage, vol. 31, n.o 3, pp. 968-980, jul. 2006, doi: 10.1016/j.neuroimage.2006.01.021.
[10] A. Schaefer et al., «Local-Global Parcellation of the Human Cerebral Cortex from Intrinsic Functional Connectivity MRI», Cereb. Cortex, vol. 28, n.o 9, pp. 3095-3114, sep. 2018, doi: 10.1093/cercor/bhx179.